變體貝葉斯高斯混合的聚集優先類型分析?
這個例子繪制了一個從toy數據集(混合了三個高斯)得到的橢球, 這些橢球是由帶有Dirichlet分布先驗的 (weight_concentration_prior_type='dirichlet_distribution') 和一個Dirichlet過程先驗(weight_concentration_prior_type='dirichlet_process')的 BayesianGaussianMixture 類模型擬合得到的。
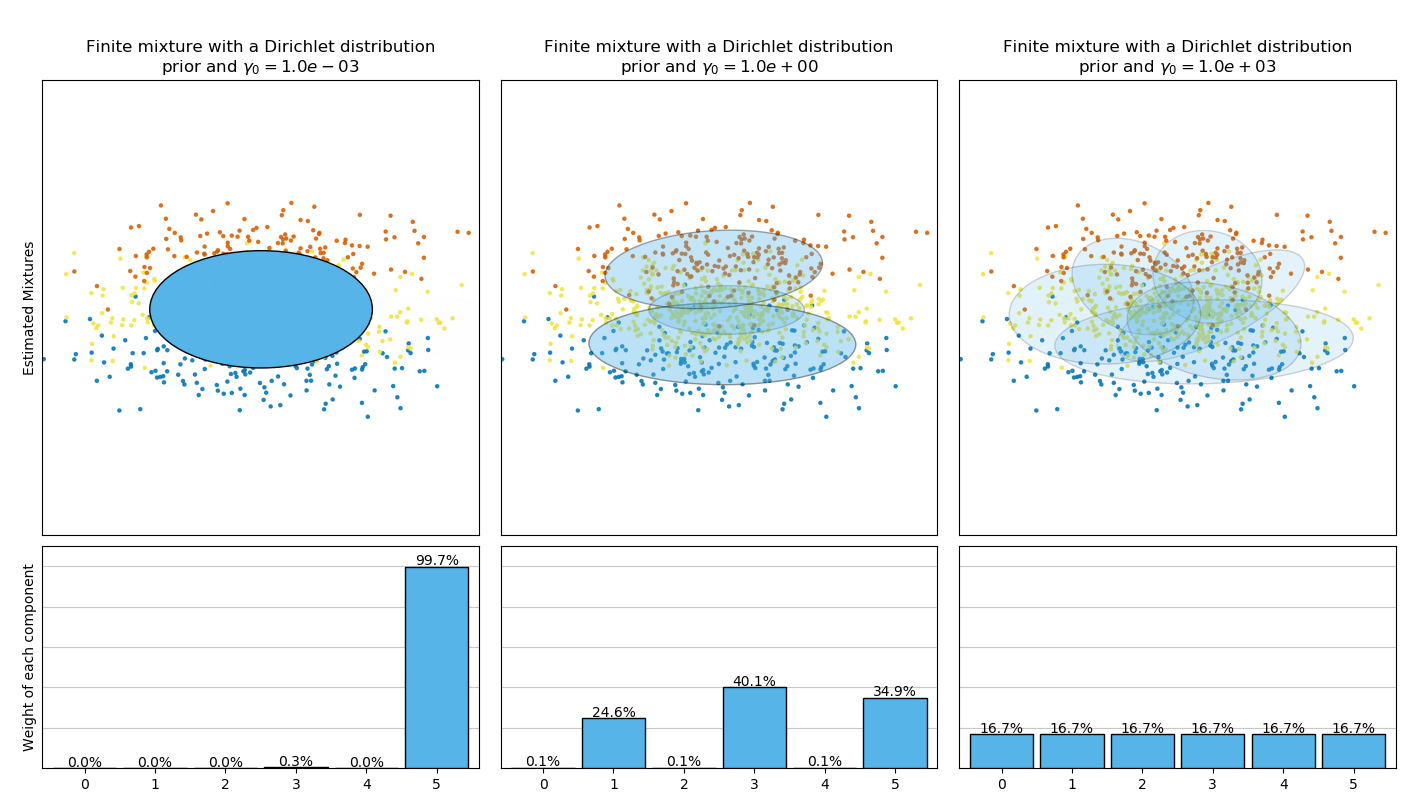
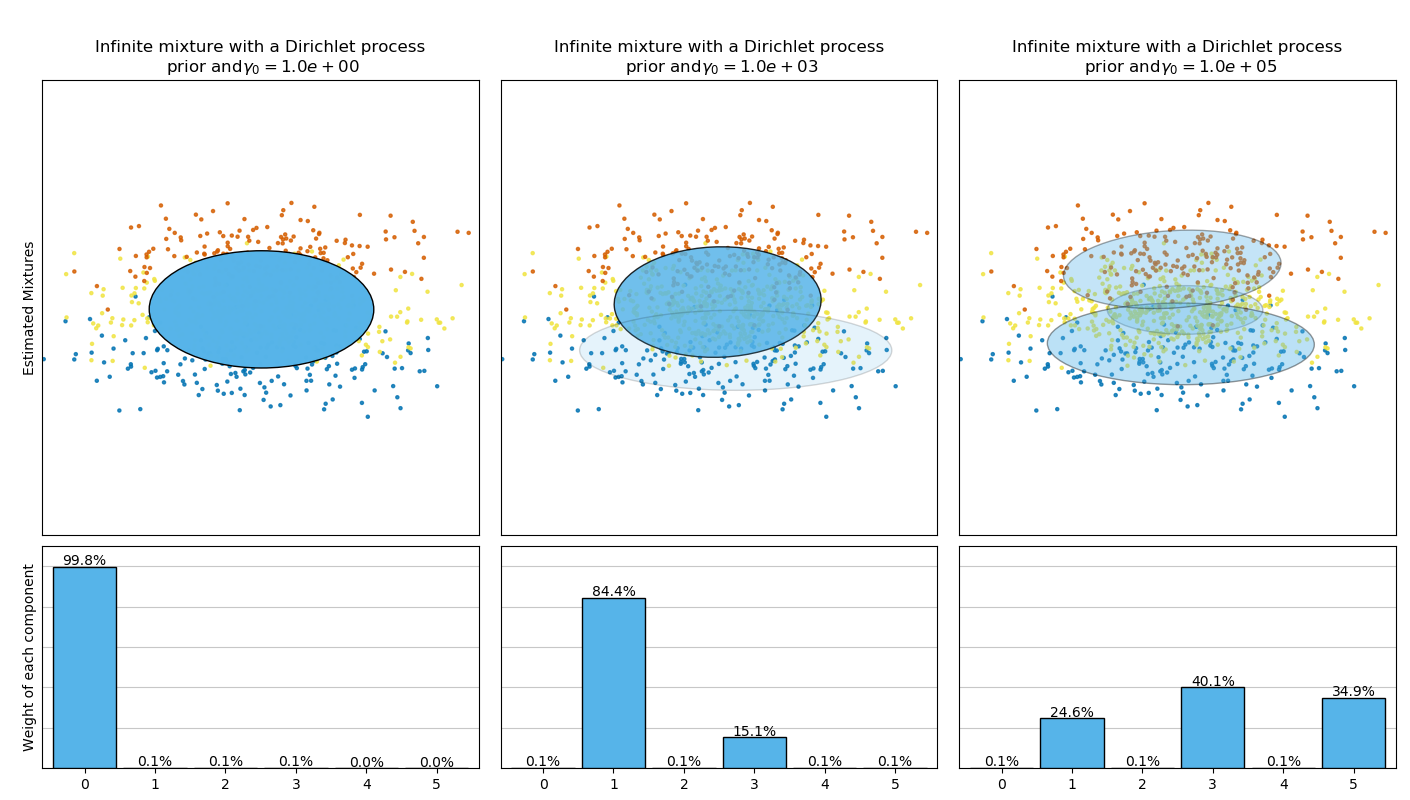
BayesianGaussianMixture類可以自動調整混合成分的個數。參數 weight_concentration_prior 與產生的具有非零權重的成分數有直接聯系。指定聚集先驗的低值將使模型將大部分權重放在少數成分上,其余成分的權重將非常接近于零。較高的聚集先驗,將允許更多的成分在混合中更活躍。
Dirichlet過程先驗允許定義無窮多個成分,并自動選擇正確的成分的個數:它只在必要時激活成分。
相反,具有Dirichlet分布先驗的經典有限混合模型更傾向于更均勻的權重成分,因此,它傾向于將自然聚類劃分為不必要的子成分。
# Author: Thierry Guillemot <thierry.guillemot.work@gmail.com>
# License: BSD 3 clause
import numpy as np
import matplotlib as mpl
import matplotlib.pyplot as plt
import matplotlib.gridspec as gridspec
from sklearn.mixture import BayesianGaussianMixture
print(__doc__)
def plot_ellipses(ax, weights, means, covars):
for n in range(means.shape[0]):
eig_vals, eig_vecs = np.linalg.eigh(covars[n])
unit_eig_vec = eig_vecs[0] / np.linalg.norm(eig_vecs[0])
angle = np.arctan2(unit_eig_vec[1], unit_eig_vec[0])
# Ellipse needs degrees
angle = 180 * angle / np.pi
# eigenvector normalization
eig_vals = 2 * np.sqrt(2) * np.sqrt(eig_vals)
ell = mpl.patches.Ellipse(means[n], eig_vals[0], eig_vals[1],
180 + angle, edgecolor='black')
ell.set_clip_box(ax.bbox)
ell.set_alpha(weights[n])
ell.set_facecolor('#56B4E9')
ax.add_artist(ell)
def plot_results(ax1, ax2, estimator, X, y, title, plot_title=False):
ax1.set_title(title)
ax1.scatter(X[:, 0], X[:, 1], s=5, marker='o', color=colors[y], alpha=0.8)
ax1.set_xlim(-2., 2.)
ax1.set_ylim(-3., 3.)
ax1.set_xticks(())
ax1.set_yticks(())
plot_ellipses(ax1, estimator.weights_, estimator.means_,
estimator.covariances_)
ax2.get_xaxis().set_tick_params(direction='out')
ax2.yaxis.grid(True, alpha=0.7)
for k, w in enumerate(estimator.weights_):
ax2.bar(k, w, width=0.9, color='#56B4E9', zorder=3,
align='center', edgecolor='black')
ax2.text(k, w + 0.007, "%.1f%%" % (w * 100.),
horizontalalignment='center')
ax2.set_xlim(-.6, 2 * n_components - .4)
ax2.set_ylim(0., 1.1)
ax2.tick_params(axis='y', which='both', left=False,
right=False, labelleft=False)
ax2.tick_params(axis='x', which='both', top=False)
if plot_title:
ax1.set_ylabel('Estimated Mixtures')
ax2.set_ylabel('Weight of each component')
# Parameters of the dataset
random_state, n_components, n_features = 2, 3, 2
colors = np.array(['#0072B2', '#F0E442', '#D55E00'])
covars = np.array([[[.7, .0], [.0, .1]],
[[.5, .0], [.0, .1]],
[[.5, .0], [.0, .1]]])
samples = np.array([200, 500, 200])
means = np.array([[.0, -.70],
[.0, .0],
[.0, .70]])
# mean_precision_prior= 0.8 to minimize the influence of the prior
estimators = [
("Finite mixture with a Dirichlet distribution\nprior and "
r"$\gamma_0=$", BayesianGaussianMixture(
weight_concentration_prior_type="dirichlet_distribution",
n_components=2 * n_components, reg_covar=0, init_params='random',
max_iter=1500, mean_precision_prior=.8,
random_state=random_state), [0.001, 1, 1000]),
("Infinite mixture with a Dirichlet process\n prior and" r"$\gamma_0=$",
BayesianGaussianMixture(
weight_concentration_prior_type="dirichlet_process",
n_components=2 * n_components, reg_covar=0, init_params='random',
max_iter=1500, mean_precision_prior=.8,
random_state=random_state), [1, 1000, 100000])]
# Generate data
rng = np.random.RandomState(random_state)
X = np.vstack([
rng.multivariate_normal(means[j], covars[j], samples[j])
for j in range(n_components)])
y = np.concatenate([np.full(samples[j], j, dtype=int)
for j in range(n_components)])
# Plot results in two different figures
for (title, estimator, concentrations_prior) in estimators:
plt.figure(figsize=(4.7 * 3, 8))
plt.subplots_adjust(bottom=.04, top=0.90, hspace=.05, wspace=.05,
left=.03, right=.99)
gs = gridspec.GridSpec(3, len(concentrations_prior))
for k, concentration in enumerate(concentrations_prior):
estimator.weight_concentration_prior = concentration
estimator.fit(X)
plot_results(plt.subplot(gs[0:2, k]), plt.subplot(gs[2, k]), estimator,
X, y, r"%s$%.1e$" % (title, concentration),
plot_title=k == 0)
plt.show()
腳本的總運行時間:(0分10.068秒)
